Scientists decode gene regulation developing human brain using machine learning
Researchers use high-throughput experiments and machine learning to identify genetic sequences affecting brain development and psychiatric disorders. Potential new treatments.
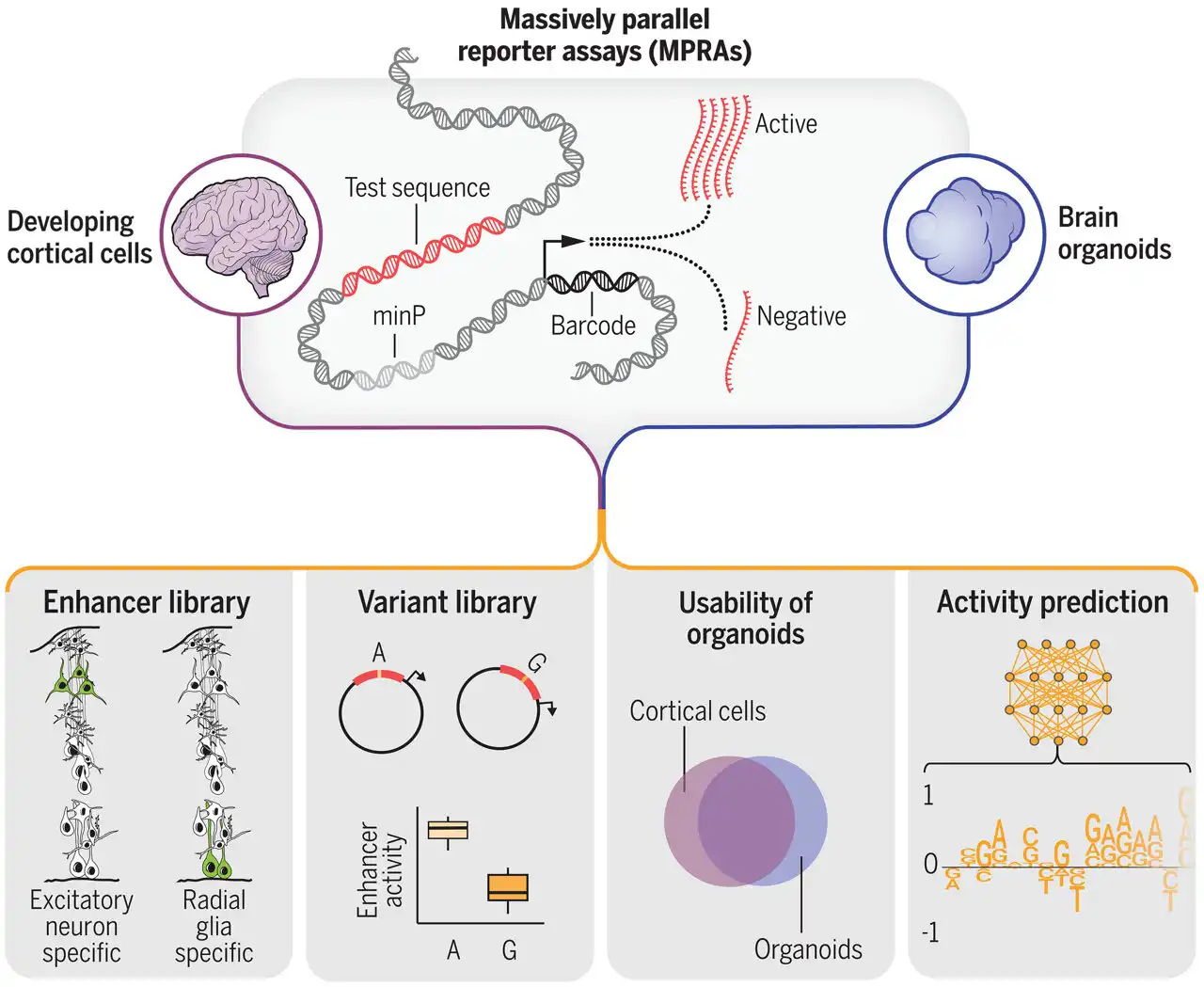
In a groundbreaking scientific achievement that expands our understanding of genetic changes influencing brain development and psychiatric disorders, a team of researchers utilized high-throughput experiments and machine learning to analyze over 100,000 sequences in human brain cells. This innovative approach led to the identification of more than 150 variants likely responsible for disease.
The collaborative study, conducted by scientists at Gladstone Institutes and the University of California, San Francisco (UCSF), has created a comprehensive catalog of genetic sequences crucial for brain development. This breakthrough paves the way for new diagnostic tools and potential treatments for neurological conditions like schizophrenia and autism spectrum disorder. Published in the journal Science, the paper titled "Massively Parallel Characterization of Regulatory Elements in the Developing Human Cortex" marks a significant milestone in the field of genetics.
Senior Investigator Katie Pollard, Ph.D., who is also the director of the Gladstone Institute for Data Science and Biotechnology, explains the extensive data collection process involving noncoding DNA regions known to play a significant role in brain function and disease. By functionally testing thousands of sequences, the research team was able to pinpoint specific genetic changes that could impact gene activity and contribute to disease development.
The study, co-led by Pollard and Nadav Ahituv, Ph.D., a professor at UCSF, also involved experimental work on brain tissue led by Tomasz Nowakowski, Ph.D. The team's findings included the identification of 164 variants linked to psychiatric disorders and over 46,000 sequences with enhancer activity in developing neurons, highlighting their role in gene regulation.
Ahituv emphasizes the potential of leveraging these enhancers to treat diseases caused by malfunctioning genes, offering a new avenue for therapeutic interventions. Furthermore, the study demonstrated the effectiveness of using brain organoids developed from human stem cells as a model for studying gene regulatory activity, with promising results that align closely with findings from real human brain tissue.
By employing machine learning techniques to predict gene activity based on DNA sequences, the research team achieved remarkable accuracy in forecasting outcomes, enabling faster and more efficient research processes. Sean Whalen, Ph.D., a senior research scientist at Gladstone and co-first author of the study, underscores the significance of this approach in accelerating discoveries and understanding the impact of genetic variants on brain cell development.
The study, conducted as part of the PsychENCODE Consortium, represents a collaborative effort to generate comprehensive gene expression and regulatory data from human brains affected by various psychiatric disorders. Through a series of studies, the consortium aims to shed light on complex conditions like autism and bipolar disorder, with the ultimate goal of revolutionizing treatment strategies.
Chengyu Deng, Ph.D., a postdoctoral researcher at UCSF and co-first author of the study, emphasizes the importance of utilizing human cells, organoids, functional screening methods, and deep learning to explore regulatory elements and genetic variants shaping human brain development. This holistic approach offers new insights into the intricate mechanisms underlying brain function and disease, paving the way for transformative advancements in the field of neuroscience.










Comments on Scientists decode gene regulation developing human brain using machine learning